Research




The alignment of natural and sexual selection


Whether natural and sexual selection reinforce or act against each other is an open question that has important implications for the evolvability of populations. In the lab we are investigating the alignment of natural and sexual selection on wings of the invasive fruit-fly Drosophila suzukii. D. suzukii has rapidly invaded many fruit-growing regions of the world (including Ontario) and causes huge economic losses to the soft-fruit industry. We are particularly interested in whether the wing shapes that are good for dispersal through flight are also favoured by females when choosing their mates. If this is the case, it might help explain the high evolvability and rapid invasion of this species throughout the world. We are addressing these questions in the lab by manipulating the phenotypes of male wings, using mate choice experiments, and assaying components of flight performance.
The evolution of sexual dimorphism
Sexual dimorphism is one of the most striking forms of phenotypic diversity observed in nature. Males and females often differ in many ways, ranging from differences in size to hormone composition to behaviour. Explaining these differences can be a challenge when we consider that males and females share almost all of their genes. For example, how can the feathers of a male bird be so bright and colourful, while those of the female which are encoded by the same genes, be brown and drab? We are interested in understanding how sexual dimorphism evolves and the genetic constraints that might prevent its evolution.
Using Drosophila as a model system, I have previously shown that the shared genome of males and females has the potential to con-strain dimorphism evolution, and that sexual dimorphism has evolved in some parts of fly wings but not in other parts.
We have ongoing projects in this area to determine whether the shared genome of males and females results in a persistent evolutionary constraint over long timescales, and to determine how sexual selection on male wings by female preferences drives the evolution of dimorphism. These are two critical and unanswered questions in evolutionary genetics and sexual selection research, respectively. Reconciling how microevolutionary processes result in macroevolutionary divergence is an ongoing challenge that we must solve in order to understand the vast biodiversity surrounding us. This work will provide critical advancement to that goal.
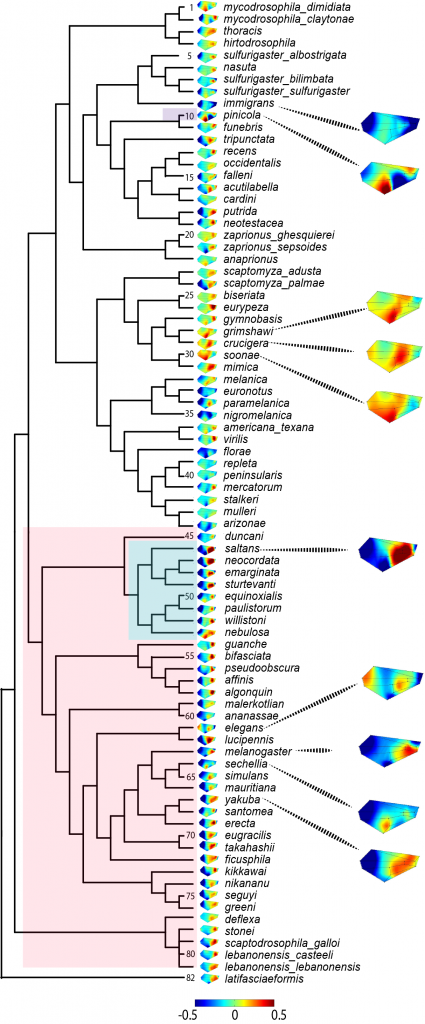
The evolution of the G matrix
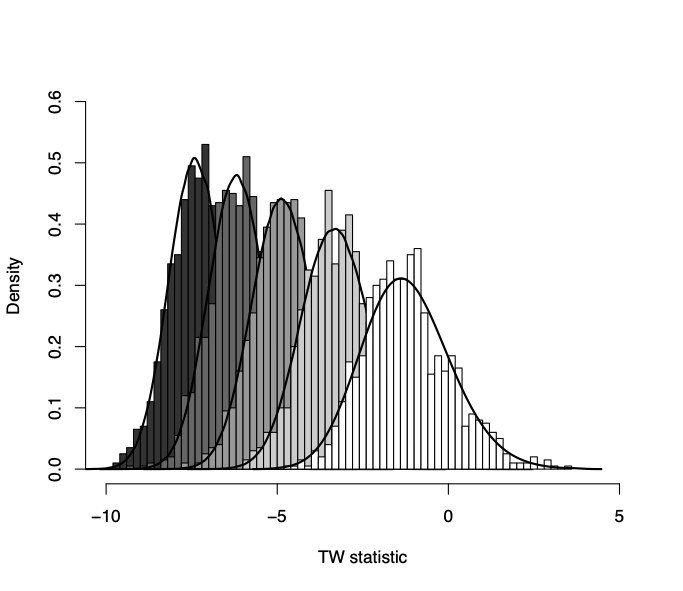
The genetic covariance matrix (G) summarizes the genetic variation in a suite of traits and the covariation between them. It is a key component of the multivariate breeder’s equation, which predicts the response to selection and thus how evolution will proceed. The breeder’s equation works pretty well at predicting short term evolution, but how well it predicts long term evolution is less clear. One reason, is that G itself can evolve in response to selection. Using Drosophila wings as a model, we are empirically investigating how G matrices of many species have diverged from each other. We are particularly interested in the timescales that G is stable for, which could indicate how long microevolutionary parameters are relevant in determining macroevolutionary change.
Along with our interest in determining empirical patterns of G matrix evolution comes the need to develop new statistical tools to study G matrices. Ongoing projects in the lab are focusing on characterizing the null distribution of genetic variance that we would expect if there were no genetic variation in G, and in which circumstances the phenotypic covariance matrix (P) is a good substitute for G. This question is highly relevant for macroevolutionary studies where P can be observed but G is rarely known.
Stabilizing selection
Stabilizing selection reduces genetic variation in a population by selecting against individuals that are extreme. Despite the key role of stabilizing selection in evolutionary theory, and its central role in determining how traits evolve across geological timescales, studies that measure selection on phenotypes in contemporary populations often find that diversifying mechanisms of selection are more prevalent. How to resolve this paradox of stasis is not clear, despite a great deal of effort.
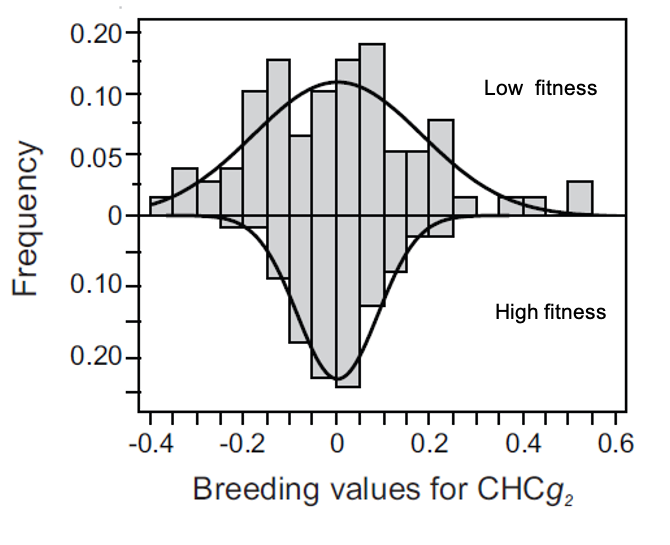
An under-appreciated aspect of stabilizing selection is that most of it should arise because of pleiotropy, where the alleles that underlie one trait also underlie many other traits. This type of stabilizing selection (called apparent stabilizing selection) is most easily uncovered by studying the genetic covariation between focal traits and fitness. Over the past five we have successfully confirmed a central role of apparent stabilising selection in the evolution of Drosophila wing and pheromone trait combinations.
What we still don’t know is how strong apparent stabilizing selection is, how the extent of pleiotropy determines its strength, and how it shapes the genomic architecture of traits. Ongoing projects in the lab aim to answer these fundamental questions, using genome wide association studies together with artificial selection experiments to characterize the genetic basis of traits with high and low genetic variation and directly link these architectures with the evolvability of populations and their ability to respond to selection.
Pleiotropy
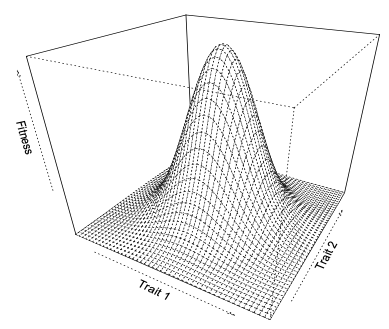
Pleiotropy is one of the key links between genotype and phenotype and occurs when one gene or mutation or allele affects more than one trait. Early models (Fisher’s geometric model) of evolution implied universal pleiotropy, where all genes affect all traits in an organism. In Fisher’s model, he likened the fitness of an organism to focusing an image on a microscope- how clear the image is (fitness) depends on the state of various knobs (phenotypes) on the microscope, and moving just one knob can throw the whole image out of focus.
The concept of universal pleiotropy fell out of favour in the early GWAS era, where scientists were preoccupied with linking specific alleles to specific phenotypes and rarely found evidence that the same SNP was significant for many traits. The problem with this approach, is that counting the number of SNPs that significantly affect one trait, and then counting how many of those significantly affect another is severely underpowered. More recently in the GWA world, an omnigenic model (a new name for universal pleiotropy) of genetic architecture has gained traction, suggesting that most genes affect most traits.
In collaboration with David Houle (Florida State University) and Thomas F. Hansen (University of Oslo), we are now taking advantage of new multivariate statistical approaches to develop “bottom-up” methods to estimate the pleiotropic effects of alleles on multiple traits with GWA data.